Slides available at: bit.ly/pybay-dupe (open as Desktop)
Jupyter Notebook source at: github.com/vintasoftware/deduplication-slides/
Introduction¶
Real world data is inputted by people and often it's:
- Not reviewed
- Not linked with related data
- Not properly normalized by the input system
- Or simply it's incorrectly inputted because people make mistakes: typos, mishearing, miscalculation, misinterpretation, etc.
This causes the following problems on data:
- lack of unique identifiers (making difficult detect duplicates in a dataset or to link with other datasets)
- duplications (e.g. multiple records refer to a single person)
- inconsistencies (e.g. a person appears with multiple addresses)
- bad formatting (e.g. birth dates appear with multiple formats like DD/MM/YY and YYYY-MM-DD)
All of that affects the ability to properly extract knowledge from one or more datasets.
The solution is to perform Record Linkage. It works by joining records in a fuzzy way using data like names, addresses, phone numbers, dates, etc.
The term Record Linkage is most used when the linkage is applied to multiple datasets, like joining a Restaurant Food Inspection dataset with an Employee Wage dataset. In fact, someone did just that.
Record Linkage is also known as Data Matching, Entity Resolution, and other names.
What we'll discuss here is a specific application of Record Linkage, called Deduplication, which is applying Record Linkage on a single dataset against itself to find which records are duplicates. Here's a very simple example:
import warnings; warnings.simplefilter('ignore')
import logging; logging.disable(level=logging.INFO)
import pandas as pd
data = [
("Chin's","3200 Las Vegas Boulevard","New York"),
("Chin Bistro","3200 Las Vegas Blvd.","New York"),
("Bistro","3400 Las Vegas Blvd.","New York City"),
("Bistro","3400 Las Vegas B.","NYC"),
]
df = pd.DataFrame(data, columns=['restaurant', 'address', 'city'])
df
A good deduplication on the data above would find that:
(0, 1)are duplicates(2, 3)are duplicates- but
(0, 1)and(2, 3)are different, despite being similar
Process¶
The process for deduplicating a dataset usually is:
- Preprocessing
- input: dataset
- output: cleaned dataset
- Indexing
- output: pairs to compare
- Comparison
- output: comparison vectors
- Classification
- output: matching/nonmatching pairs
- Clustering
- output: unique record clusters
Let's explore each of those steps!
0/4 - Preprocessing¶
Without unique identifiers, we need to match records by fuzzy data like:
- Names
- Addresses
- Phone Numbers
- Dates
So it's important to clean them for matching.
Cleaning names (companies or people)¶
Let's use string functions and regexes to normalize names and remove undesired variations:
import numpy as np
import re
import pprint
company_names = company_names_dirty = [
'APPLE COMPUTER INC',
'APPLE COMPUTER, INC.',
'APPLE INC',
'Apple Computer',
'Apple Computer Co.',
'Apple Computer Company',
'Apple Computer Inc',
'Apple Computer Incorporated',
'Apple Computer, Inc.',
'Apple Inc',
'Apple Inc.',
'Apple, Inc.'
]
print("Lower case:")
company_names = [c.lower() for c in company_names]
pprint.pprint(company_names)
print("Remove irrelevant separators:")
irrelevant_regex = re.compile(r'[^a-z0-9\s]')
company_names = [irrelevant_regex.sub(' ', c) for c in company_names]
pprint.pprint(company_names)
print("Remove multi-spaces:")
multispace_regex = re.compile(r'\s\s+')
company_names = [multispace_regex.sub(' ', c).strip() for c in company_names]
pprint.pprint(company_names)
print("Remove stopwords:")
business_stopwords = { # suppose we got this from somewhere
'computer',
'co',
'company',
'corp',
'corporation',
'inc',
'incorporated',
'llc',
#...
}
company_names = [
' '.join([c_part for c_part in c.split() if c_part not in business_stopwords])
for c in company_names
]
pprint.pprint(company_names)
We can also use the natural language processing library probablepeople to parse company names and extract just the parts we want (or break into parts and match by part on the comparison step later):
import probablepeople as pp
pp.parse("Apple Computer Incorporated")
company_names_alternative_1 = [
[
parsed_value
for parsed_value, parsed_type
in pp.parse(c)
if parsed_type == 'CorporationName'
]
for c in company_names_dirty
]
pprint.pprint(company_names_alternative_1)
probablepeople, as the name suggests, can parse people names too:
pp.parse('Mr. Guido van Rossum')
But performance isn't great from non-English names. So you can try nameparser, which is not probabilistic:
from nameparser import HumanName
from IPython.display import display
display(pp.parse('Flávio Juvenal da Silva Jr'))
display(HumanName('Flávio Juvenal da Silva Jr'))
If it's useful to ignore accents, try unidecode:
import unidecode
print("ASCII transliteration:")
brazilian_name = "Flávio"
print(brazilian_name, "->")
print(unidecode.unidecode(brazilian_name))
It's important to consider how the data values were inputted. If the value was inputted by someone while hearing from someone else, try phonetic encoding with doublemetaphone to normalize words that sound similar:
from doublemetaphone import doublemetaphone
print(doublemetaphone('Andrew'))
print(doublemetaphone('André'))
If data was obtained via Optical Character Recognition (OCR), techniques exist to correct common misspellings that OCRs make (like C to L, S to 5, q to g, etc). See Christen, page 47 [2].
Cleaning addresses¶
Geocoding street addresses, i.e., converting them to latitude/longitude is very useful for matching, because geocoders usually clean irrelevant address variations. Also, having lat/lng enables the calculation of geometric distances between addresses.
import requests
import geocoder
full_addresses = [
"2066 Crist Drive, 94024, Los Altos, CALIFORNIA, US",
"2066 Crist Dr, 94024, LOS ALTOS, CALIFORNIA, US",
"20863 Stevens Creek Blvd., Suite 300, 95015, CUPERTINO, CALIFORNIA, US",
"20863 STEVENS CREEK BLVD STE 330, 95014, Lupertino, CALIFORNIA, US",
"10260 Bandley Drive, 95014, Cupertino, CALIFORNIA, US",
"10260 Bandley Dr., 95014, Cupertino, CALIFORNIA, US",
"20525 MARIANI AVENUE, 95014, CUPERTINO, CALIFORNIA, US",
"20525 Mariani Ave, 95014, CUPERTINO, CALIFORNIA, US",
"1 Infinite Loop, 95014, Cupertino, CALIFORNIA, US",
"One Infinite Loop,, 95014, Cupertino, CALIFORNIA, US",
"One Apple Park Way, 95014, Cupertino, CALIFORNIA, US",
"1 Apple Park Way, 95014, Cupertino, CALIFORNIA, US",
]
full_addresses_latlng = []
with requests.Session() as session:
for a in full_addresses:
a_geocoded = geocoder.google(a, session=session)
full_addresses_latlng.append(a_geocoded.latlng)
address_latlng = list(zip(full_addresses, full_addresses_latlng))
pprint.pprint((address_latlng[2], address_latlng[3]))
print()
Google geocoder is able to:
- Ignore lower/upper case difference
- Ignore the difference between Suite 300 vs 330 (building entrance may be the same)
- Consider 'Lupertino' as 'Cupertino'
- Expand abbreviations, like 'STE' vs 'Suite'
pprint.pprint((address_latlng[8], address_latlng[9]))
However, it gets slighly different addresses on the case of '1' vs 'One'.
Note geocoding from web APIs is slow and has quota limits! Alternatively, you can build your own geocoder with:
If geocoding is inviable, we can use the library pypostal to normalize addresses:
from postal.expand import expand_address
print(full_addresses[8])
pprint.pprint(expand_address(full_addresses[8]))
print()
print(full_addresses[9])
pprint.pprint(expand_address(full_addresses[9]))
print(full_addresses[2])
pprint.pprint(expand_address(full_addresses[2]))
print()
print(full_addresses[3])
pprint.pprint(expand_address(full_addresses[3]))
import usaddress
print(full_addresses[0])
pprint.pprint(usaddress.parse(full_addresses[0]))
print()
print(full_addresses[1])
pprint.pprint(usaddress.parse(full_addresses[1]))
Cleaning phone numbers¶
phonenumbers library can normalize phone numbers from many countries:
import phonenumbers
print("Phone number normalization:")
phone = "(541) 555-3010"
print(phone, '->')
print(
phonenumbers.format_number(
phonenumbers.parse(phone, 'US'),
phonenumbers.PhoneNumberFormat.E164)
)
Cleaning dates¶
dateparser library can guess date formats and parse them as datetime objects. It can even guess DD/MM or MM/DD by the language:
import dateparser
print(dateparser.parse("at 10/1/2018 10am"))
print(dateparser.parse("às 10/1/2018 10:00"))
pprint.pprint(dateparser.DateDataParser().get_date_data("às 10/1/2018 10:00"))
Check also dateutil parser with fuzzy=True.
Cleaning a real dataset¶
Now we know the tools, let's grab a dataset to preprocess and go on the other deduplication steps. Our dataset is based* on the Restaurant dataset, a well-known dataset used by researchers. It contains 881 restaurant records and contains 150 duplicates.
* We've introduced some changes that you can check by doing a diff restaurant.original.csv restaurant.csv
df_with_truth = pd.read_csv('restaurant.csv', skip_blank_lines=True)
df_with_truth.head(9)
The dataset comes with the true matches indicated by the 'cluster' column. We'll remove it. We'll also remove the 'phone' to makes things more difficult:
df = df_with_truth.drop(columns=['cluster', 'phone'])
df.head(9)
By running info method, we assert our dataset is quite clean, missing only 1 value of 'type'. We'll just replace it with an empty string. Note that on other datasets you might need to do more profiling to know what to fix. Check Mull's talk [1].
df.info(verbose=True, null_counts=True)
df = df.fillna('')
Now we'll clean the column values! Cleaning name:
def assign_no_symbols_name(df):
return df.assign(
name=df['name']
.str.replace(irrelevant_regex, ' ')
.str.replace(multispace_regex, ' '))
df = assign_no_symbols_name(df)
df.head(9)
from collections import Counter
possible_stopwords = Counter(" ".join(df["name"]).split()).most_common(20)
pprint.pprint(possible_stopwords)
def assign_cleaned_name(df):
restaurant_stopwords = {
's', 'the', 'la', 'le', 'of', 'and', 'on', 'l'}
restaurant_stopwords_regex = r'\b(?:{})\b'.format(
'|'.join(restaurant_stopwords))
return df.assign(
name=df['name']
.str.replace(restaurant_stopwords_regex, '')
.str.replace(multispace_regex, ' ')
.str.strip())
df = assign_cleaned_name(df)
df.head(9)
Geocoding addr:
all_addresses = df['addr'].str.cat(df['city'], sep=', ').values
unique_addresses = np.unique(all_addresses)
print(len(all_addresses), len(unique_addresses))
import os.path
import json
geocoding_filename = 'address_to_geocoding.json'
def geocode_addresses(address_to_geocoding):
remaining_addresses = (
set(unique_addresses) -
set(k for k, v in address_to_geocoding.items() if v is not None))
with requests.Session() as session:
for i, address in enumerate(remaining_addresses):
print(f"Geocoding {i + 1}/{len(remaining_addresses)}")
geocode_result = geocoder.google(address, session=session)
address_to_geocoding[address] = geocode_result.json
with open(geocoding_filename, 'w') as f:
json.dump(address_to_geocoding, f, indent=4)
if not os.path.exists(geocoding_filename):
address_to_geocoding = {}
geocode_addresses(address_to_geocoding)
else:
with open(geocoding_filename) as f:
address_to_geocoding = json.load(f)
geocode_addresses(address_to_geocoding)
address_to_postal = {
k: v['postal']
for k, v in address_to_geocoding.items()
if v is not None and 'postal' in v
}
address_to_latlng = {
k: (v['lat'], v['lng'])
for k, v in address_to_geocoding.items()
if v is not None
}
print(f"Failed to get postal from {len(address_to_geocoding) - len(address_to_postal)}")
print(f"Failed to get latlng from {len(address_to_geocoding) - len(address_to_latlng)}")
def assign_postal_lat_lng(df):
addresses = df['addr'].str.cat(df['city'], sep=', ')
addresses_to_postal = [address_to_postal.get(a) for a in addresses]
addresses_to_lat = [address_to_latlng[a][0] if a in address_to_latlng else None for a in addresses]
addresses_to_lng = [address_to_latlng[a][1] if a in address_to_latlng else None for a in addresses]
return df.assign(postal=addresses_to_postal, lat=addresses_to_lat, lng=addresses_to_lng)
df = assign_postal_lat_lng(df)
df.head(6)
Adding addr_variations:
def assign_addr_variations(df):
return df.assign(
addr_variations=df['addr'].apply(
lambda addr: frozenset(expand_address(addr))))
df = assign_addr_variations(df)
df[['name', 'addr', 'addr_variations']].head(6)
Now, with a clean dataset, we can move to the next step.
1/4 - Indexing¶
To explain the following deduplication steps, we'll use the Python libary recordlinkage, aka Python Record Linkage Toolkit. We choose it because it doesn't abstract too much the details of the process, even though it has a simple API.
We have the cleaned records, we now need the pairs we want to compare to find matches. To produce the pairs, we could do a "full" index, i.e., all records against all records:
import recordlinkage as rl
full_indexer = rl.FullIndex()
pairs = full_indexer.index(df)
print(f"Full index: {len(df)} records, {len(pairs)} pairs")
The formula for the total number of pairs is:
len(df) * (len(df) - 1) / 2 == 387640

The number of pairs grows too fast as the number of records grows: it grows quadratically. That's why we need indexing. We need to produce only pairs that are good candidates of being duplicates to avoid wasting too much time.
Indexing is also called blocking because the trivial way to index is to produce pairs that have some column value in common. By doing this, we produce blocks of record pairs and compare only those in the same block:

Below we produce pairs that have equal values for postal code:
postal_indexer = rl.BlockIndex('postal')
postal_index_pairs = postal_indexer.index(df)
print(f"Postal index: {len(postal_index_pairs)} pairs")
We could also produce pairs by sorting the unique values of some column and, for each value, getting the records with neighboring values. The idea is to produce pairs with close values on some column, like johnny and john or 2018-01-02 and 2018-01-05. It looks like this:

Below we produce pairs that have neighboring values for name:
name_indexer = rl.SortedNeighbourhoodIndex('name', window=7)
name_index_pairs = name_indexer.index(df)
print(f"Name index: {len(name_index_pairs)} pairs")
Note that simply sorting values wouldn't be able to get kamila and camila as neighbors, as sorting is sensitive to the leading characters of strings. There are other ways to index that could handle that, check Christen, chapter 4 [2].
To produce more pairs without introducing redundant pairs, we should union different indexes:
pairs = postal_index_pairs.union(name_index_pairs)
print(f"Postal or name index: {len(pairs)} pairs")
We now have which pairs we want to run comparisons on!
2/4 - Comparison¶
Now we want to run comparisons on the indexed pairs to produce a comparison vector for each pair. A comparison vector represents the similarity between 2 records by holding similarity values between 0 to 1 for each column.
pd.DataFrame([[1.0, 0.75, 0.0, 0.25, 0.0]],
columns=['name', 'addr', 'type', 'latlng', 'addr_variations'],
index=pd.MultiIndex.from_arrays([[100], [200]]))
For example, the comparison vector above means the pair of records (100, 200) has:
- Equal
names - Similar
addrs - Completely different
types - Some distance on
latlngs - No agreement on
addr_variations
To compute the comparison vectors for all indexed pairs, we define a similarity function for each column:
vectorized_intersection = np.vectorize(
lambda x, y: float(bool(x.intersection(y))))
def compare_addr_variations(a1, a2):
return vectorized_intersection(a1, a2)
comp = rl.Compare()
comp.string('name', 'name', method='jarowinkler', label='name')
comp.string('addr', 'addr', method='jarowinkler', label='addr')
comp.string('city', 'city', method='jarowinkler', label='city')
comp.string('type', 'type', method='jarowinkler', label='type')
comp.geo('lat', 'lng', 'lat', 'lng', method='exp', scale=0.5, label='latlng')
comp.compare_vectorized(compare_addr_variations,
'addr_variations', 'addr_variations',
label='addr_variations');
There are many similarity functions we could use for strings. For example:
jarowinklergives priority to the begining of the stringlevenshteincares more about the orderlcscares less about the order
from recordlinkage.algorithms.string import (
jarowinkler_similarity as jarowinkler,
levenshtein_similarity as levenshtein,
longest_common_substring_similarity as lcs)
for s1, s2 in [["arnie morton", "arnie morton s of chicago"],
["the palm", "palm the"]]:
print(f'jarowinkler("{s1}", "{s2}") ==',
jarowinkler([s1], [s2])[0])
print(f'levenshtein("{s1}", "{s2}") ==',
levenshtein([s1], [s2])[0])
print(f'lcs("{s1}", "{s2}") ==',
lcs([s1], [s2])[0])
print()
For more details on the different similarity functions you can use, check Christen, chapter 5 [2]. Also, check the similarity functions implemented on recordlikage library.
Now we'll compute the comparison vectors:
comparison_vectors = comp.compute(pairs, df)
comparison_vectors.head(5)
It's good to check the statistics of the vectors to see if they look right:
comparison_vectors.describe()
By looking at the summary we can see it's easier to get higher similarities on 'city', 'addr' and 'type', not so easy on 'name' and 'latlng', and very difficult on 'addr_variations'. Therefore, it seems that:
- we have few unique values of 'city' in our dataset and that also causes...
- ...lots of 'addr' to be similar
- there only a limited number of 'type's, so it repeats a lot
- and getting a 1 on 'addr_variations' is rare since it only considers an exact agreement on address variation
Now, with our comparison vectors, we'll explore different ways to classify them as matches and nonmatches.
3/4 - Classification¶
Before, let's remember what's on records 0 to 5:
df.head(6)
Threshold-Based Classification¶
A simple way to classify comparison vectors as matches or nonmatches is to compute a weighted average over the vectors to get a score:
scores = np.average(
comparison_vectors.values,
axis=1,
weights=[30, 10, 5, 10, 30, 15])
scored_comparison_vectors = comparison_vectors.assign(score=scores)
scored_comparison_vectors.head(5)
By looking at the data, we know record 0 truly matches 1 and 2, so we'll classify as a match any pair with score >= 0.85. That's our threshold:
matches = comparison_vectors[
scored_comparison_vectors['score'] >= 0.85]
matches.head(5)
Let's check another match we've found with this threshold: what are the matches of record 3?
display(df.loc[[3]])
print("matched:")
display(df.loc[matches.loc[(3,)].index])
Seems OK! Since we have the true match status on cluster column, we can evaluate how well our threshold classification did:
golden_pairs = rl.BlockIndex('cluster').index(df_with_truth)
print("Golden pairs:", len(golden_pairs))
found_pairs_set = set(matches.index)
golden_pairs_set = set(golden_pairs)
true_positives = golden_pairs_set & found_pairs_set
false_positives = found_pairs_set - golden_pairs_set
false_negatives = golden_pairs_set - found_pairs_set
print('true_positives total:', len(true_positives))
print('false_positives total:', len(false_positives))
print('false_negatives total:', len(false_negatives))
We've got a small number of false positives. Some are really tricky cases:
print(f"False positives:")
for false_positive_pair in false_positives:
display(df.loc[list(false_positive_pair)][['name', 'addr', 'city', 'type', 'lat', 'lng']])
On the other hand, we got a lot of false negatives. We've missed a lot of matches!
print(f"False negatives (sample 10 of {len(false_negatives)}):")
for false_negative_pair in list(false_negatives)[:10]:
display(df.loc[list(false_negative_pair)][['name', 'addr', 'city', 'type', 'lat', 'lng']])
We've set the weights and the threshold by guessing, could we do any better?
Supervised Classification¶
Instead of trying to guess weights and thresholds, we can train a classifier to learn how to classify matches and nonmatches based on some training data we provide:
df_training = pd.read_csv('restaurant-training.csv', skip_blank_lines=True)
df_training = df_training.drop(columns=['phone'])
df_training
We need to preprocess our training data too:
df_training = assign_no_symbols_name(df_training)
df_training = assign_cleaned_name(df_training)
df_training = assign_postal_lat_lng(df_training)
df_training = assign_addr_variations(df_training)
df_training.head(5)
We'll feed a Support Vector Machine classifier with our training data. SVMs are resilient to noise, can handle correlated features (like 'addr' and 'latlng') and are robust to imbalanced training sets. That last attribute is relevant for deduplication, because we'll usually find more negative pairs than positive pairs to add to a training set. (Bilenko [3])
all_training_pairs = rl.FullIndex().index(df_training)
matches_training_pairs = rl.BlockIndex('cluster').index(df_training)
training_vectors = comp.compute(all_training_pairs, df_training)
svm = rl.SVMClassifier()
svm.learn(training_vectors, matches_training_pairs);
svm_pairs = svm.predict(comparison_vectors)
svm_found_pairs_set = set(svm_pairs)
svm_true_positives = golden_pairs_set & svm_found_pairs_set
svm_false_positives = svm_found_pairs_set - golden_pairs_set
svm_false_negatives = golden_pairs_set - svm_found_pairs_set
print('true_positives total:', len(true_positives))
print('false_positives total:', len(false_positives))
print('false_negatives total:', len(false_negatives))
print()
print('svm_true_positives total:', len(svm_true_positives))
print('svm_false_positives total:', len(svm_false_positives))
print('svm_false_negatives total:', len(svm_false_negatives))
Much better results! The only false positive we got on the SVM classifier and not on the threshold method is a really difficult case where most columns are very similar:
print("(SVM false positives) - (Threshold false positives):")
for svm_false_positive in (svm_false_positives - false_positives):
display(df.loc[list(svm_false_positive)][['name', 'addr', 'city', 'type', 'lat', 'lng']])
But the SVM was tricked by some apparently simple cases, so we can't be very confident it really learned well to classify matches:
print("(SVM false negatives) - (Threshold false negatives):")
for svm_false_negative in (svm_false_negatives - false_negatives):
display(df.loc[list(svm_false_negative)][['name', 'addr', 'city', 'type', 'lat', 'lng']])
There are other classifiers from recordlinkage library we could try, but the truth is:
- It's very difficult to build a good training set that takes in account all important cases of matches/nonmatches
- It's possible to tune classifier parameters to get better results, but it's very difficult to decide the right parameters that will generalize well for future predictions
- And we're not even sure if the indexing rules we used are really sane: we can be dropping true positives that are not being blocked together, or even introducing false negatives that are being blocked together but our classifier isn't being able classify them as nonmatching
The alternative to all that uncertainty is...
Active Learning Classification¶
Active Learning methods identify training examples that "lead to maximal accuracy improvements" (Bilenko [3]), both to train optimal classifier weights, as well as to find optimal indexing/blocking rules! A Python library called Dedupe implements that.
We'll see it in practice. First we define the fields/columns of our data:
import logging; logging.disable(level=logging.NOTSET)
import dedupe
fields = [
{
'field': 'name',
'variable name': 'name',
'type': 'ShortString',
'has missing': True
},
{
'field': 'addr',
'variable name': 'addr',
'type': 'ShortString',
},
{
'field': 'city',
'variable name': 'city',
'type': 'ShortString',
'has missing': True
},
] # ...
Dedupe comes with built-in variables for common data types like names, addresses, latlong, datetimes, thereby reducing the preprocessing work. It also supports custom variables:
def addr_variations_comparator(x, y):
return 1.0 - float(bool(x.intersection(y)))
fields.extend([
{
'field': 'postal',
'variable name': 'postal',
'type': 'ShortString',
'has missing': True
},
{
'field': 'latlng',
'variable name': 'latlng',
'type': 'LatLong',
'has missing': True
},
{
'field': 'addr_variations',
'variable name': 'addr_variations',
'type': 'Custom',
'comparator': addr_variations_comparator
},
])
Other nice feature of Dedupe is the built-in ability to model interactions) between fields. What's that for? Imagine we have an exact address match: it makes no sense to have all 'addr', 'city', 'postal', 'latlng', 'addr_variations' contributing additively to the matching odds. It's expected all of them to match if 'addr' and 'city' match exactly. So to prevent a redundant increase of odds, we should add a interaction on those fields:
fields.extend([
{
'type': 'Interaction',
'interaction variables': [
'addr',
'city',
'postal',
'latlng',
'addr_variations'
]
}
])
Now we initialize the Dedupe instance with the fields. We can also use a pre-saved pickled Dedupe instance:
settings_filename = 'dedupe-settings.pickle'
if os.path.exists(settings_filename):
with open(settings_filename, 'rb') as sf:
deduper = dedupe.StaticDedupe(sf, num_cores=4)
else:
deduper = dedupe.Dedupe(fields, num_cores=4)
We need to adapt the data a bit to the format Dedupe wants:
data_for_dedupe = df.to_dict('index')
for record in data_for_dedupe.values():
# Change nans to None
for k, v in record.items():
if isinstance(v, float) and np.isnan(v):
record[k] = None
# Move lat and lng to a single field latlng
lat = record.pop('lat')
lng = record.pop('lng')
if lat is not None and lng is not None:
record['latlng'] = (lat, lng)
else:
record['latlng'] = None
Here we're using a Dedupe instance that we trained before. Let's check how was the training input/output:
training_input_output = 'training-input-output.txt'
if os.path.exists(training_input_output):
with open(training_input_output) as t:
txt = t.read()
print('\n'.join(txt.split('\n')[:114]))
print('...')
You can check the full training at training-input-output.txt.
If you want to train it yourself, do a rm dedupe-settings.pickle dedupe-slides-training.json and run this whole Active Learning session again.
if not isinstance(deduper, dedupe.StaticDedupe):
deduper.sample(data_for_dedupe)
training_filename = 'dedupe-slides-training.json'
if os.path.exists(training_filename):
with open(training_filename) as tf:
deduper.readTraining(tf)
dedupe.consoleLabel(deduper)
with open(training_filename, 'w') as tf:
deduper.writeTraining(tf)
deduper.train()
with open(settings_filename, 'wb') as sf:
deduper.writeSettings(sf)
After training, we can see which blocking predicates (indexing rules) the deduper learned from our training input. It's good to do that to check if we trained enough:
deduper.predicates
Those predicates make sense:
- Pairs where the first token of the name is the same AND the addr is similar
- ... union with ...
- Pairs where the name shares two tokens AND the fingerprint of the name is the same (fingerprint means the sorted tokens)
The deduper selected those predicates from this extense list of possible predicates:
deduper.data_model.predicates()
To proceed with the deduplication, we compute the clustering threshold and call the actual match:
import itertools
threshold = deduper.threshold(data_for_dedupe, recall_weight=2)
clustered_dupes = deduper.match(data_for_dedupe, threshold)
dedupe_found_pairs_set = set()
for cluster, __ in clustered_dupes: # we'll explain that later
for pair in itertools.combinations(cluster, 2):
dedupe_found_pairs_set.add(tuple(pair))
Now we'll evaluate how it performed:
dedupe_true_positives = golden_pairs_set & dedupe_found_pairs_set
dedupe_false_positives = dedupe_found_pairs_set - golden_pairs_set
dedupe_false_negatives = golden_pairs_set - dedupe_found_pairs_set
print('true_positives total:', len(true_positives))
print('false_positives total:', len(false_positives))
print('false_negatives total:', len(false_negatives))
print()
print('svm_true_positives total:', len(svm_true_positives))
print('svm_false_positives total:', len(svm_false_positives))
print('svm_false_negatives total:', len(svm_false_negatives))
print()
print('dedupe_true_positives total:', len(dedupe_true_positives))
print('dedupe_false_positives total:', len(dedupe_false_positives))
print('dedupe_false_negatives total:', len(dedupe_false_negatives))
Great! It found more true positives than the other methods. But it also found more false positives than SVM. It's possible to control that by lowering recall_weight:
threshold_low_recall = deduper.threshold(data_for_dedupe, recall_weight=0.5)
clustered_dupes_low_recall = deduper.match(data_for_dedupe, threshold_low_recall)
dedupe_low_recall_found_pairs_set = set()
for cluster, __ in clustered_dupes_low_recall:
for pair in itertools.combinations(cluster, 2):
dedupe_low_recall_found_pairs_set.add(tuple(pair))
dedupe_low_recall_true_positives = golden_pairs_set & dedupe_low_recall_found_pairs_set
dedupe_low_recall_false_positives = dedupe_low_recall_found_pairs_set - golden_pairs_set
dedupe_low_recall_false_negatives = golden_pairs_set - dedupe_low_recall_found_pairs_set
print('dedupe_true_positives total:', len(dedupe_true_positives))
print('dedupe_false_positives total:', len(dedupe_false_positives))
print('dedupe_false_negatives total:', len(dedupe_false_negatives))
print()
print('dedupe_low_recall_true_positives total:', len(dedupe_low_recall_true_positives))
print('dedupe_low_recall_false_positives total:', len(dedupe_low_recall_false_positives))
print('dedupe_low_recall_false_negatives total:', len(dedupe_low_recall_false_negatives))
As we can see, there's always a tradeoff between true positives and false positives, or, more precisely, between precision and recall. You have to decide what's more important for your business.
But let's suppose we want to find more true positives and use the previous dedupe_found_pairs_set. What false positives it found?
print("Dedupe false positives")
for false_positive_pair in list(dedupe_false_positives):
display(df.loc[list(false_positive_pair)][['name', 'addr', 'city', 'type', 'lat', 'lng']])
The deduper is confused by "caffe"s that are located near each other. "caffe" could be a stopword? Or even better, we could incorporate some logic for name rarity: if the token isn't rare, like "caffe", it should contribute less with the name similarity than a rarer token like "reggio". It's possible to do that with an interaction with name frequency.
Or maybe the deduper could have learned a more restrictive name blocking? Or the classifier should have put less weight on name? It's difficult to guess because those things could have introduced more false negatives too. What about them?
print("Dedupe false negatives")
for false_negative_pair in list(dedupe_false_negatives):
display(df.loc[list(false_negative_pair)][['name', 'addr', 'city', 'type', 'lat', 'lng']])
Some of these false negatives could be prevented with better address normalization, but on others, the data is simply bad: different addresses for the same place, maybe a corner, maybe two entrances? We could try to train more the deduper to fix this. But we'll leave as it is and move to the last step of the deduplication process.
4/4 - Clustering¶
Using the Threshold or the SVM, we got the matching pairs. But what Dedupe returned to us were clusters of matches:
clustered_dupes[:5]
Dedupe went one step further on the process and merged the matching pairs into clusters! Why is that important? Because the following can happen:
- We have the records A, B, and C
- By deduplicating, we found the matching pairs (A, B) and (B, C). However, (A, C) was found to be a nonmatch
It doesn't make sense to consider (A, B) and (B, C) as a match, but (A, C) as a nonmatch. The solution for that ambiguity is computing the Transitive Closure with Clustering.
Using some private methods, it's possible to get the unclustered pairs from Dedupe:
from dedupe.core import scoreDuplicates
candidate_records = itertools.chain.from_iterable(deduper._blockedPairs(deduper._blockData(data_for_dedupe)))
dedupe_matches = scoreDuplicates(candidate_records,
deduper.data_model,
deduper.classifier,
deduper.num_cores)
dedupe_unclustered_found_pairs_set = {tuple(pair) for ([*pair], __) in dedupe_matches}
Let's evaluate those unclustered pairs against the clustered pairs:
dedupe_unclustered_true_positives = golden_pairs_set & dedupe_unclustered_found_pairs_set
dedupe_unclustered_false_positives = dedupe_unclustered_found_pairs_set - golden_pairs_set
dedupe_unclustered_false_negatives = golden_pairs_set - dedupe_unclustered_found_pairs_set
print('dedupe_true_positives total:', len(dedupe_true_positives))
print('dedupe_false_positives total:', len(dedupe_false_positives))
print('dedupe_false_negatives total:', len(dedupe_false_negatives))
print()
print('dedupe_unclustered_true_positives total:', len(dedupe_unclustered_true_positives))
print('dedupe_unclustered_false_positives total:', len(dedupe_unclustered_false_positives))
print('dedupe_unclustered_false_negatives total:', len(dedupe_unclustered_false_negatives))
We've found the unclustered pairs are different from the clustered pairs! That means the clustering process can both create new matches and drop found matches. Therefore, even though clustering is necessary to disambiguate the deduplication result, it can either improve or worsen the quality of found pairs.
diff_set = dedupe_found_pairs_set ^ dedupe_unclustered_found_pairs_set
display(diff_set)
Here's a case where the clustering process got a new correct match:
from graph_utils import show_cluster_graphs
good_diff_all_ids = {34, 35, 36, 37}
show_cluster_graphs(
df,
golden_pairs_set, dedupe_found_pairs_set, dedupe_unclustered_found_pairs_set,
good_diff_all_ids)
And here's a case where the clustering process dropped a true match:
bad_diff_all_ids = {6, 7, 8}
show_cluster_graphs(
df,
golden_pairs_set, dedupe_found_pairs_set, dedupe_unclustered_found_pairs_set,
bad_diff_all_ids)
Remember the recall_weight parameter of the deduper.threshold method? That's what it controls: how aggressive we want to be on finding or dropping matches while clustering. Check Dedupe docs for more details on how it performs clustering and how it computes a good threshold.
Finally, it's worth mentioning there's a web-based product version of Dedupe. If you don't want to write code for deduping a dataset, check it.
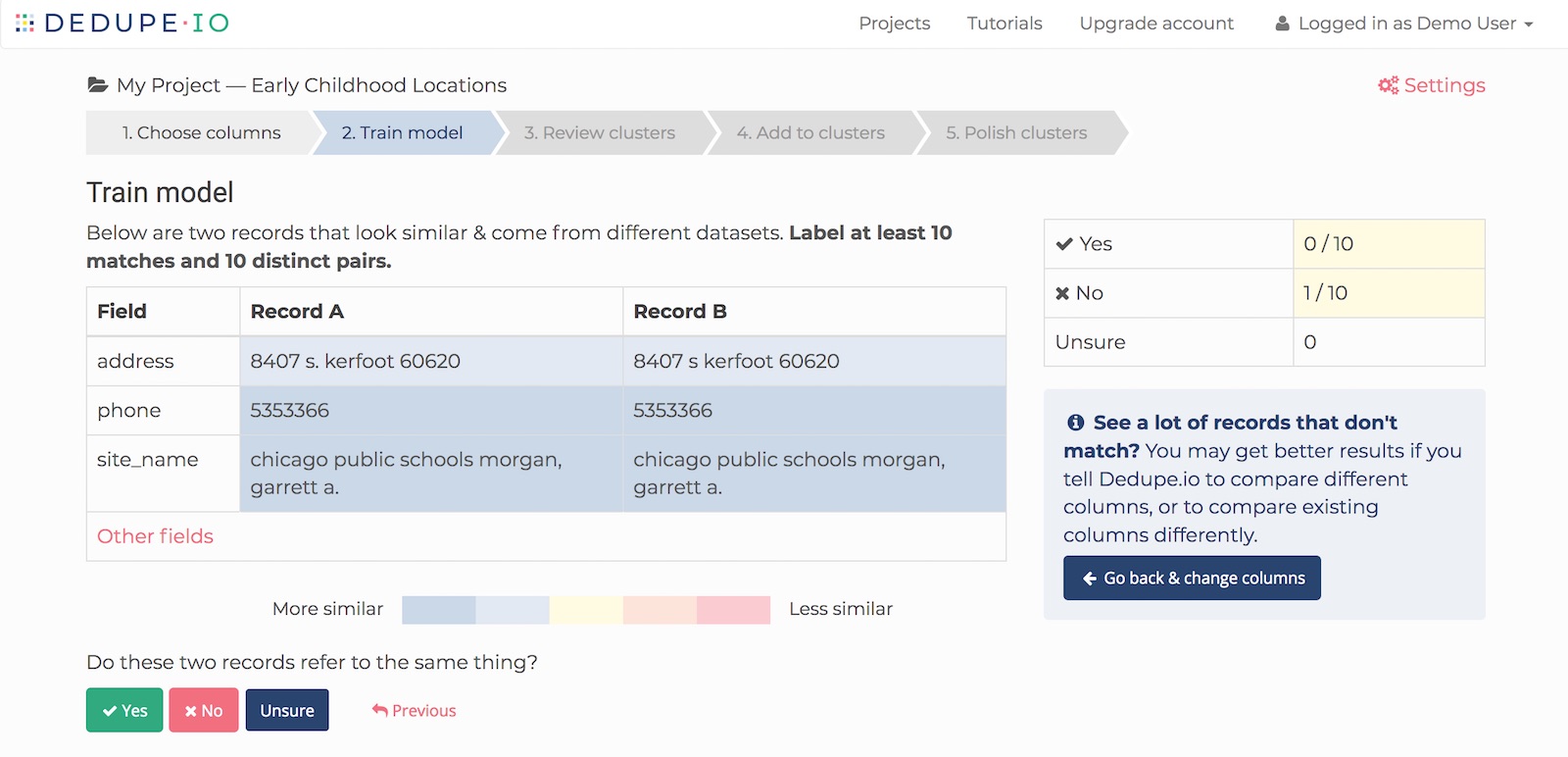
Next Steps¶
Once we have the clusters, how to consolidate data from many records into one? Check for material on Data Fusion:
- Open issue at Python Record Linkage Toolkit
- Christen, 6.12 Merging Matches [2]
What if new records arrive? Should we merge, unmerge, move records from clusters? Check for material on Incremental Record Linkage:
- Dedupe approach is to either add to an existing cluster or create a new cluster. You can use a Gazetteer with previously deduped data, then merge new matches into it, and index new nonmatches.
- Other approaches, check papers:
Also worth checking the Privacy implications of Record Linkage:
- Christen, chapter 8 [2]
- k-anonymity, k-map, δ-presence
- Why differential privacy is awesome
References¶
- Referenced by this talk:
- Other talks about Record Linkage with Python:
- Basics of Entity Resolution with Python and Dedupe
- Automating your Data Cleanup with Python
- Comparison of Data Matching software
- Compilation of Record Linkage resources
Thank you!¶
flavio@vinta.com.br
@flaviojuvenal
vinta.com.br
Special thanks to Russell Keith-Magee @freakboy3742, Forest Timothy Gregg @forestgregg, and Jonathan de Bruin @J535D165.
